New research explores a genetic link for high methane-producing sheep by analysing data on gut microbial populations with the animal’s genomic information, and measurements of the amount of methane it produces through burping.
Dr Elizabeth Ross applied this approach to dairy cattle in 2013, predicting high and low methane emitters from rumen microbial profiles – the first time anyone had used microbial populations to predict the host phenotype.
An accurate DNA prediction test for methane production could help farmers produce the same amount of meat and wool from their livestock with a lessened environmental impact.
Researchers have used genomics to accurately predict a sheep’s methane output levels.
The work forms part of a multi-disciplinary Australian project to reduce the contribution of livestock to greenhouse gas emissions.
Research fellow at the Queensland Alliance for Agriculture and Food Innovation, Dr Elizabeth Ross, studied sheep methane data collated by the University of New England, in conjunction with genomic markers from the sheep’s gut microbiomes to predict methane output.
Genomic prediction is the prediction of total genetic value using genome-wide dense marker maps.
The technology was pioneered by Elizabeth’s supervisor, Professor Ben Hayes, Director of the Centre for Animal Science. It is now widely used in livestock and crops to predict future trait outcomes, and is increasingly used in human disease research.
Dr Ross said researchers wanted to be able to tell which sheep produce high levels or low levels of methane, which is a greenhouse gas and also causes a loss of energy from the animal’s system.
“Energy lost as methane means that the animals are not using that same energy to grow and produce,” she said.
“We used a genomics approach to look at what different species of microbes are in the rumen and also how abundant each one is. “
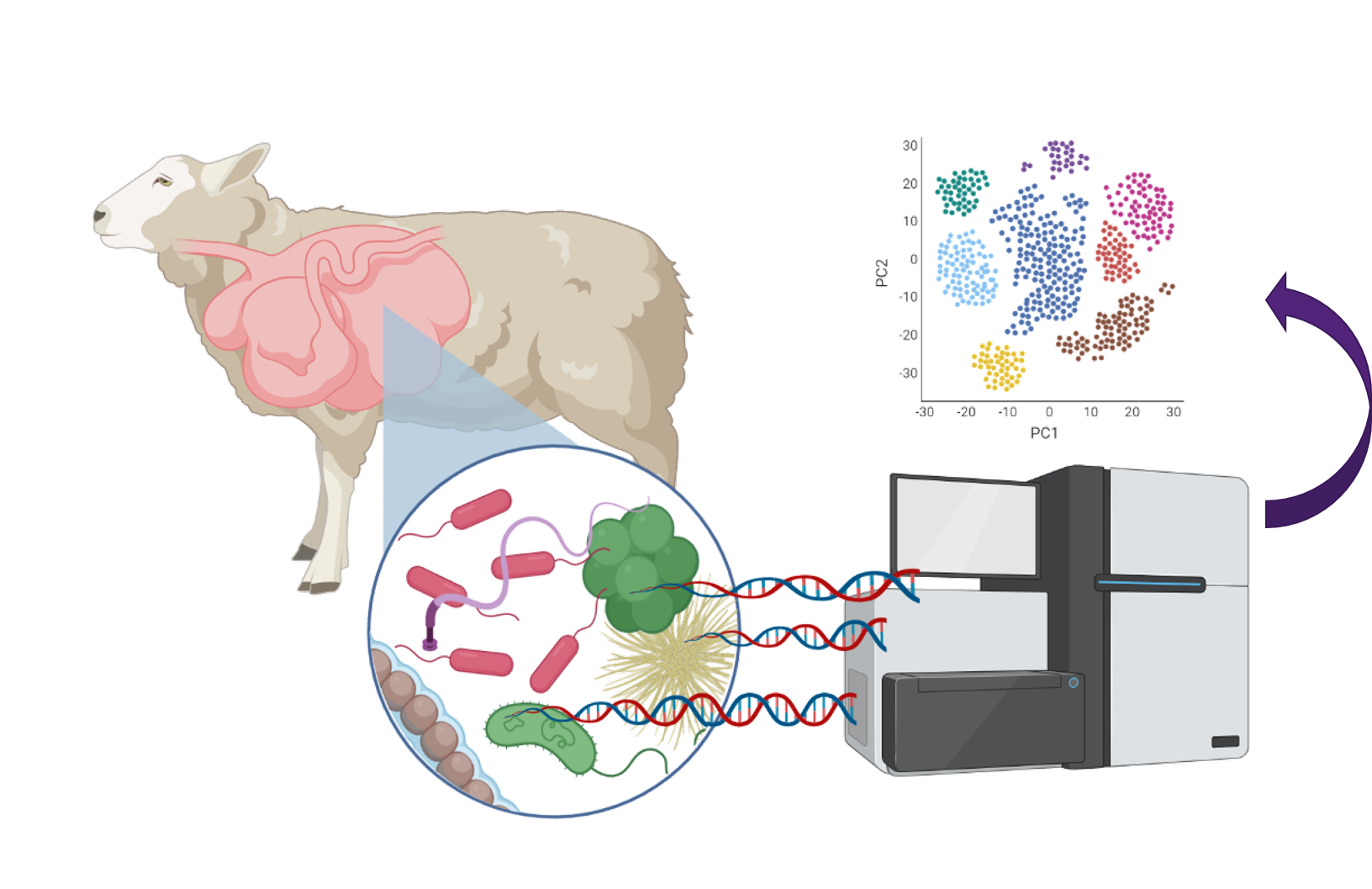
This approach looks at the entire gut microbial population in the sheep’s rumen and then compares different animals to see if those populations are very similar or very different.
This information is used in conjunction with genomic markers from across the animal’s genome, to predict how much methane that animal is going to produce.
Dr Ross was able to accurately predict methane output from a population of 99 Merino-cross sheep, based on differences in the genome.
“When we look at the different microbes and the chemicals those microbes produce in the rumen, we can use them to predict which animals will produce high or low levels of methane,” she said.
“The work begins to integrate -omics data into prediction methodologies and apply its predictive ability to increase our understanding of methane output.
“Integrating genomic, metabolomic and microbiome relationships to predict enteric methane production showed higher accuracy than either method alone.”
Dr Ross used data from a parent study conducted by University of New England researcher Victor Hutton Oddy, who collated methane emission data from the 99 sheep using a respiration chamber.
The aim is to create tools that could help farmers produce the same amount of meat and wool from their livestock with a lessened environmental impact.
Dr Ross says the work will potentially be used to not only reduce greenhouse gas emissions from Australian livestock – estimated at 71 per cent of total emissions created by the agricultural sector – but also to inform farm practices so that more is produced from the same inputs.
The work was part of the ‘Host control of methane emissions from sheep’ project, one of four projects coordinated Ruminant Pangenome Project, a national program led by Professor Phil Vercoe at the University of Western Australia (UWA).
The sheep were in a structured population of half-siblings in four families (lambs from four different sires).
They were studied over a three-year period and their microbiomes and methane output measured at two different time points.
Dr Ross said when sheep had similar microbial populations, they produced similar amounts of methane.
“But sheep’s microbial populations can be quite different because it can depend on things like how quickly food moves through the rumen and different things that might be excreted through, for example, the saliva,” she said.
“There is also always the very big effect of diet. If you change the diet, you change the microbes that are growing in the rumen.
“You can manipulate the microbial population in that way – so if you feed animals a diet that reduces methane, the microbes that grow in the rumen will have a similar population structure to a naturally low methane-emitting animal.”
“If we can identify which animals within a breed naturally produce less methane, we can use those,” Dr Ross said.
“That means that for that breed, without compromising any production metrics, we can lower methane emissions because we’ll know which ones are naturally producing less methane.
“Those individuals are the ones we want to use because they’re producing just as much meat as the other animals in the flock but less methane. So if we breed from them we can pass on the genetic component.”
Dr Ross says a long-term goal of the research is to help farmers identify which sheep are naturally low methane emitters, so they can make informed decisions about which animals they have on their property.
“There is an inverse relationship between feed efficiency, where animals are converting the energy in their feed into products such as muscle or milk or wool, versus releasing it as methane.
“That energy can either go into the animal or it can go out of the animal in the form of methane and is lost. It potentially has a really nice advantage where animals can be more efficient and also better for the environment.”
She says the methane production predictions were about 20 to 30 per cent accurate, meaning its 20 per cent of the way to a perfect prediction.
“That is on the lower side but it’s significant in the sense that it was a real difference that we were picking up and it allowed us to identify low methane emitters. What we hope is to be able to get bigger numbers of animals so that we can improve that accuracy.”
The project was a collaboration of researchers from the Queensland Alliance for Agriculture and Food Innovation, the New South Wales Department of Primary Industries, UNE, and CSIRO and UWA.
The work was funded by Meat and Livestock Australia the Department of Agriculture, Fisheries and Forestry under the Filling the Research Gap program.
Contact: Dr Elizabeth Ross, Research Fellow, Centre for Animal Science, E: e.ross@uq.edu.au, T: +61 7 334 62162
QAAFI Communications: E: qaaficomms@uq.edu.au, T: +61 (0)7 3346 2092, M: +61 439 399 886
The Queensland Alliance for Agriculture and Food Innovation is a research institute at The University of Queensland, established with and supported by the Queensland Department of Primary Industries.




