Queensland researchers have discovered that a previously unrecognised bacterium is responsible for the signs of lung disease found in pig carcases, rather than a similar, internationally recognised infection that the animals had been vaccinated against.
Having solved the disease mystery, the research team at the Queensland Alliance for Agriculture and Food Innovation (QAAFI) is now working to develop on-farm tests and treatments for the new infection.
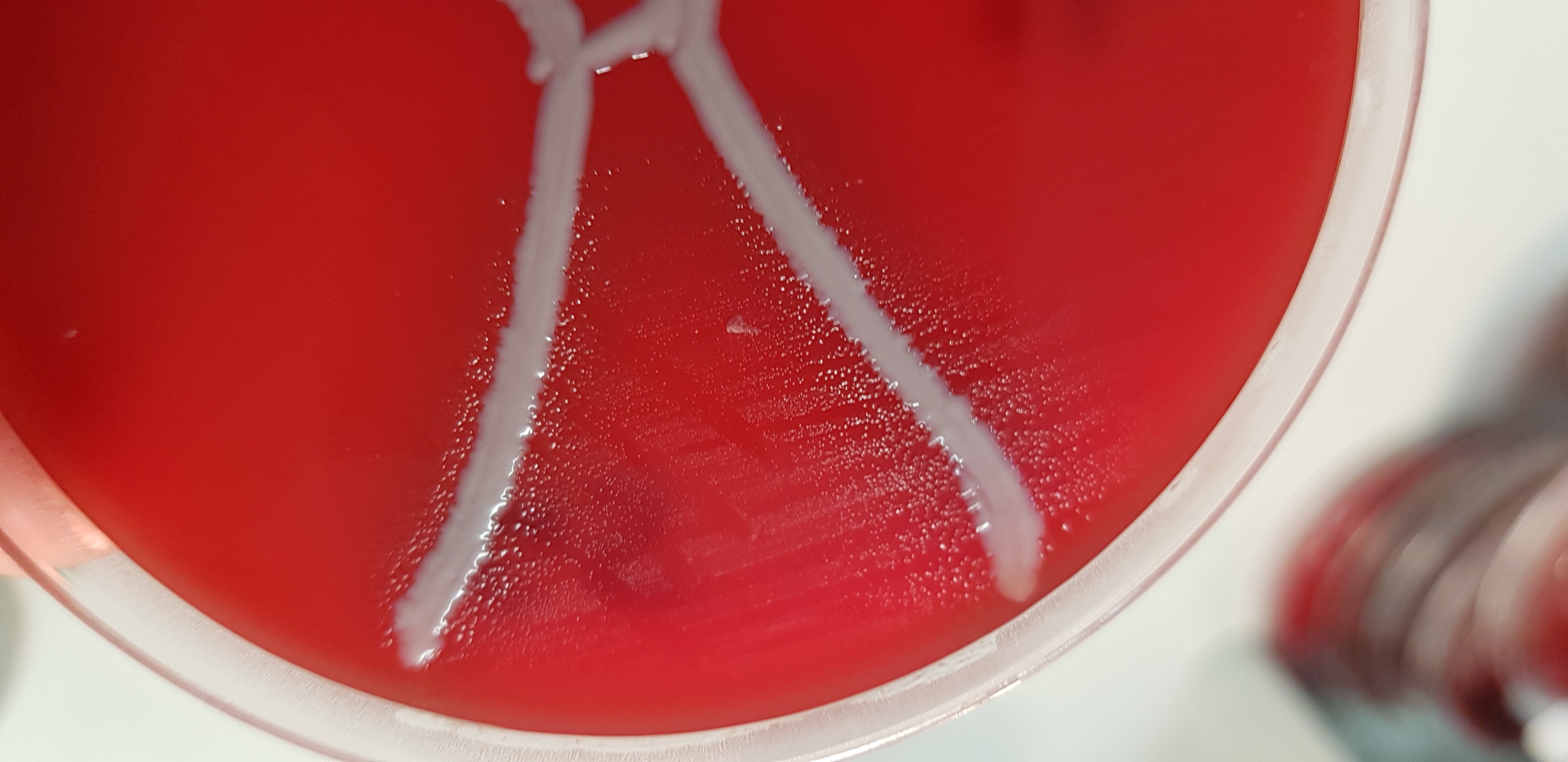
Several years ago it was noticed that lesions, abscesses and pleurisy found in the lungs of pigs at abattoirs looked very similar to those associated with a known serious pig respiratory disease, porcine pleuropneumonia. This is caused by the bacterium Actinobacillus pleuropneumoniae, which was assumed to be the culprit, despite the fact that animals had had been fully vaccinated.
Porcine pleuropneumonia is a major economic disease that causes animals to lose weight at a critical growth stage. Previous research has shown that animals’ average daily gain can drop by up to 20% until halted by treatment, with the animals requiring an extra 20 or so days to recover. This leads to a considerable increase in production costs, or if a producer decided to sell the animals underweight, the losses could be as high as $60 per pig.
New species discovered
Australian Pork Limited put QAAFI researchers on the case. They discovered one new species and another potential new species of lung-infecting bacteria, which put to rest concerns that current vaccines simply weren’t working.
Project leader Dr Conny Turni from the University of Queensland says that when the unexplained signs of disease were found, it was in the same growth period in which porcine pleuropneumonia caused by Actinobacillus pleuropneumoniae occurs.
However, the pigs are vaccinated for this so there could only be two possibilities: either the vaccines weren’t working, or there was another pathogen at work causing a similar disease or in some way interfering with the efficacy of the vaccines.
“We had been storing isolates from some diseased pigs but hadn’t been able to identify them until a couple of years ago when we had two master’s students work on them, and they determined that a number of these isolates represented a new bacterial species,” says Dr Turni.
However, the researchers couldn’t continue the formal process of describing and naming the new species because the discovery occurred at the same time that the closest known relative to the new organism, Haemophilus parasuis, was being renamed Glaesserella by US researchers. Once the new genus was formally recognised in 2019, the QAAFI researchers could announce Glaesserella australis as a new species.
On-farm effects
Dr Turni says G. australis is associated with two disease scenarios. One is where there are no apparent clinical signs of disease on-farm, but at the abattoir the carcase has lesions and abscesses in the lungs that are very similar to those caused by Actinobacillus pleuropneumoniae.
In the other disease scenario G. australis causes clinical signs in pigs on-farm, at 12 to 20 weeks of age, with some cases being fatal.
Dr Turni says continuing research into G. australis has led to a diagnostic assay that is currently being validated. This involves testing 26 G. australis isolates, 15 reference strains and one field isolate of A. pleuropneumoniae, 16 reference strains for another bacterium (Pasteurella multocida) that causes respiratory disease, ,and another 47 strains and field isolates representing 12 genera and 26 species of similar bacteria.
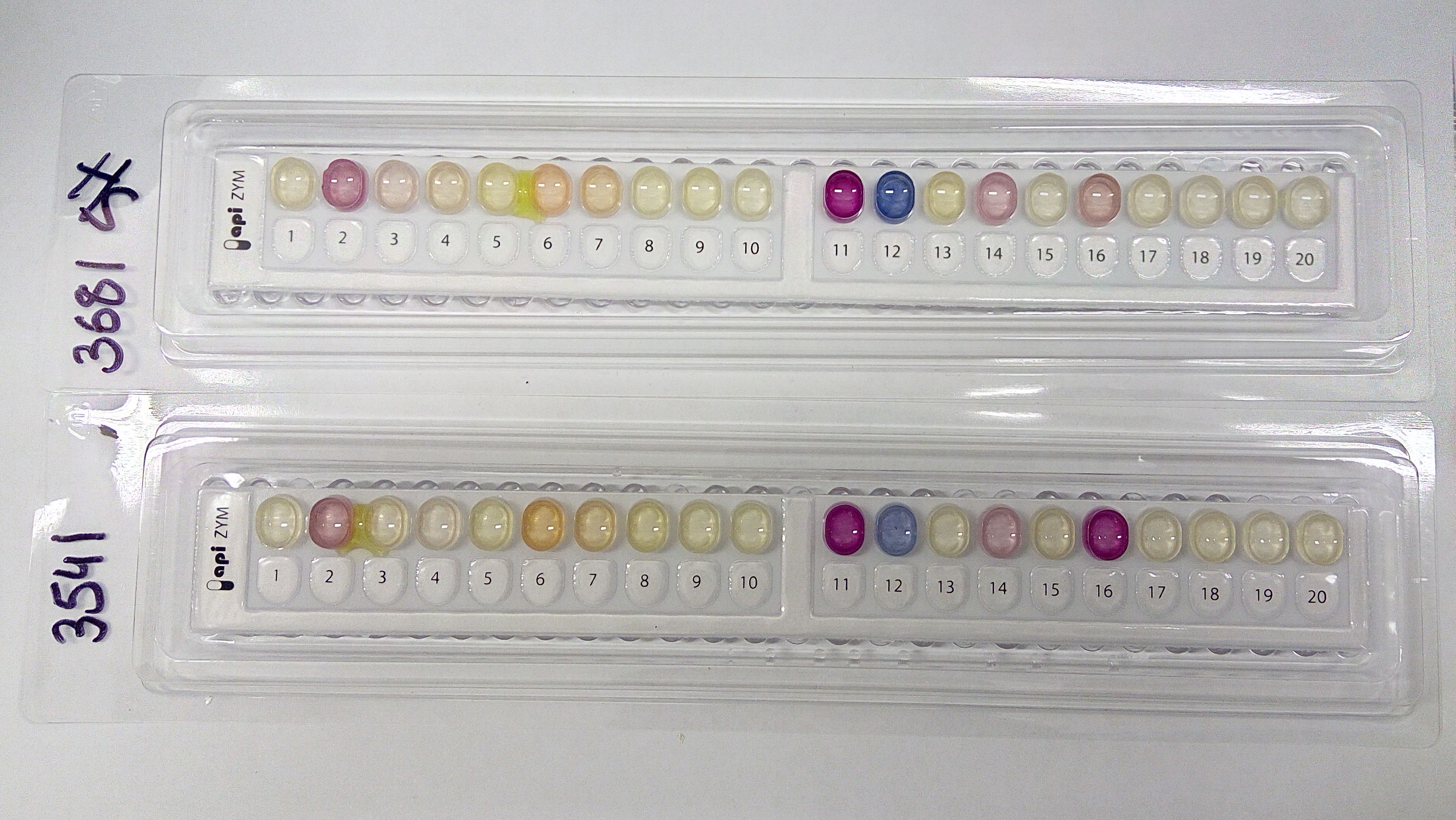
To determine the prevalence of G. australis, the researchers sampled lungs with lesions, abscesses and pleurisy from 23 farms in NSW, 43 in Queensland, one in SA and 27 in Victoria. This data is still being analysed.
For future on-farm diagnosis, the QAAFI team is investigating the potential for nasal and tonsil swabbing to see if this will detect the bacterium in live pigs, which would simplify control and management of the disease.
Another part of the project is examining methods to determine the antimicrobial sensitivity profile of G. australis isolates to help the industry develop targeted, effective treatment programs.
Manager, Production Innovation at Pork Australia, Rebecca Athorn says the discovery of G. australis delivers both animal health and economic benefits for the industry.
“The judicious use of antimicrobials and antibiotics is an industry priority. This discovery, and the development of an assay to test for this particular bug, allowing targeted treatment, is significant to the industry’s efforts overall to increase vaccine efficiency,” she says.
This research is funded by Australian Pork Limited and the Department of Agriculture and Fisheries.
Contact: Dr Conny Turni, Senior Research Fellow, Centre for Animal Science, Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, T: +61 7 344 32463, M: 0432 548 199 or E: c.turni1@uq.edu.au
High-resolution photos: For more photos and captions contact Carolyn Martin QAAFI Communications, M: 0439 399 886 E: carolyn.martin@uq.edu.au
Related articles:
Lesions, abscesses and pleurisy - Australian scientists identify bacteria causing lung infection in pigs - ABC Rural report by Jennifer Nichols published 6 May 2020.
Breathtaking research led by a quiet achiever - Australian Pork Newspaper by Brendon Cant published 1 May 2020.
The Queensland Alliance for Agriculture and Food Innovation is a research institute at The University of Queensland, established with and supported by the Queensland Department of Primary Industries.



